Main scientific focus areas:
1. Deciphering the chromatin regulatory functions of SIRT6 and SIRT7 and the ticking of the “Epigenetic Clock”: We pioneered the discovery that these sirtuins are highly selective histone deacetylases. Our work revealed that SIRT6 targets H3K9ac and H3K56ac, while SIRT7 deacetylates H3K18ac and H3K36ac—modifications central to gene regulation, DNA repair, and heterochromatin maintenance.
Currently, our lab is investigating how these modifications alter the rate of biological aging by altering the “ticking of the DNA Methylation Clock”. This biomarker serves as the best known biomarker for biological age and has an especially tight link to histone modifications at H3K36, and likely H3K36ac.
2. Understanding how sirtuin dysfunction drives cellular senescence and aging: We uncovered novel triggers of senescence, including pathologic transcription from pericentric heterochromatin (regulated by SIRT6) and ribosomal DNA instability within nucleoli (guarded by SIRT7). These discoveries reveal fundamental mechanisms linking chromatin dysfunction to cellular aging.
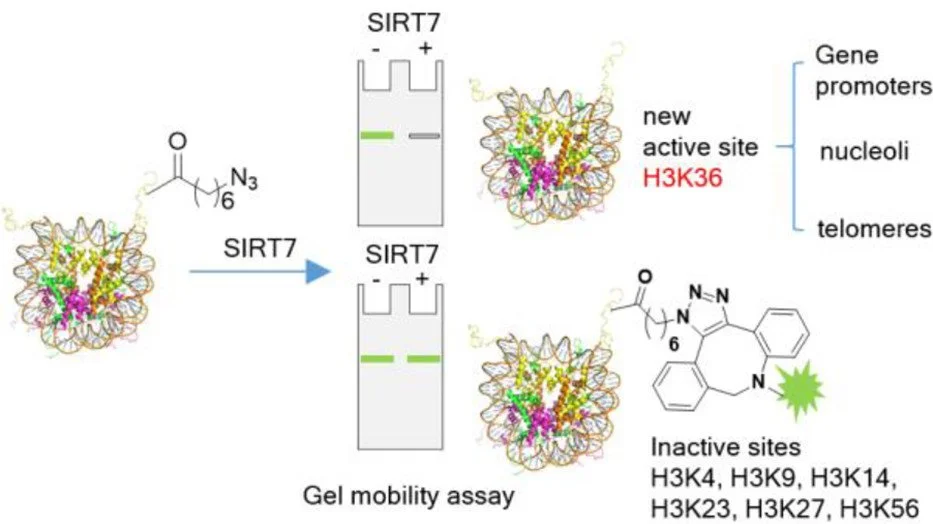
3. Elucidating the molecular circuitry of sirtuin action: Through proteomic approaches, we identify novel substrates, protein interactions, and post-translational modifications that regulate SIRT6 and SIRT7 function. This includes our recent discovery of a novel physiologic substrate for SIRT7 (H3K36ac) and dynamic SIRT6 phosphorylation during mitosis and stress responses.
4. Exploring sirtuin functions in cancer and metabolic disease: We demonstrated that SIRT7 maintains oncogenic transformation through H3K18 deacetylation, while SIRT6 acts as a tumor suppressor. Both sirtuins also regulate metabolic homeostasis, with implications for fatty liver disease, obesity, and diabetes, which we are currently characterizing using new mouse models established in our lab.
5. Developing strategies for therapeutic intervention: By understanding how sirtuins coordinate multiple protective pathways, we aim to identify selective approaches to modulate their activity for treating age-related diseases while avoiding potential adverse effects.
The dynamic language of Lysine Acetylation
Post-translational modifications (PTMs) exponentially expand the functional diversity of the proteome, with lysine acetylation serving as a critical regulatory mechanism in cellular physiology. The acetylation reaction involves the addition of an acetyl group to the ε-amino group of lysine residues, neutralizing their positive charge and fundamentally altering protein structure and function.
This seemingly simple chemical modification orchestrates complex biological processes through a sophisticated enzymatic machinery. Lysine acetyltransferases (KATs) catalyze acetyl group addition, while histone deacetylases (HDACs) remove these modifications, creating a dynamic regulatory system that responds to diverse cellular signals.
Lysine acetylation reaction: The chemical structure of lysine and acetylated derivatives. Lysine acetylation is performed KATs, which replace the hydrogen moiety on the side chain with an acetyl group. Acetyl groups can be removed by HDACs.
Histone (De)Acetylation: A Master Regulator of Chromatin
Within the nucleus, histone acetylation serves as a primary mechanism for regulating chromatin accessibility and gene expression. The neutralization of lysine's positive charge weakens histone-DNA interactions, promoting chromatin opening and transcriptional activation. Conversely, deacetylation typically leads to chromatin compaction and gene silencing.
This acetylation-deacetylation cycle is remarkably dynamic, with modifications being added and removed on timescales of minutes to hours—far more rapidly than many other histone modifications. This dynamism allows cells to rapidly respond to environmental cues, stress signals, and metabolic changes.
Sirtuins: NAD+-Dependent Guardians of Cellular Homeostasis
Among the diverse HDAC families, sirtuins (Class III HDACs) stand apart due to their unique dependence on NAD+ as a cofactor. This requirement directly links their enzymatic activity to the cell's metabolic and energetic state, positioning Sirtuins as critical sensors that couple metabolism to epigenetic regulation.
Unlike zinc-dependent HDACs, sirtuins consume NAD+ during catalysis, producing nicotinamide and O-acetyl-ADP-ribose. This mechanism enables sirtuins to function as metabolic sensors, with their activity rising and falling with cellular NAD+ levels—a feature that becomes particularly important during aging, stress, and disease states where NAD+ metabolism is altered.
Mammalian sirtuins exhibit remarkable substrate specificity. While classical HDACs often show promiscuous activity, SIRT6 and SIRT7 demonstrate exquisite selectivity for specific histone residues and chromatin contexts. This selectivity allows for precise regulation of distinct genomic regions and cellular pathways, making them attractive targets for therapeutic intervention.
Nuclear Sirtuins: SIRT6 and SIRT7
Our laboratory focuses on the two predominantly nuclear mammalian sirtuins—SIRT6 and SIRT7—which function as highly selective histone deacetylases with profound impacts on aging and disease.
SIRT6: The Guardian of Genome Stability
SIRT6 emerged from our research as a critical regulator linking chromatin dynamics to fundamental aging processes. We discovered that SIRT6 selectively deacetylates H3K9ac and H3K56ac, establishing it as the first mammalian sirtuin with robust histone deacetylase activity.
Key Functions and Mechanisms:
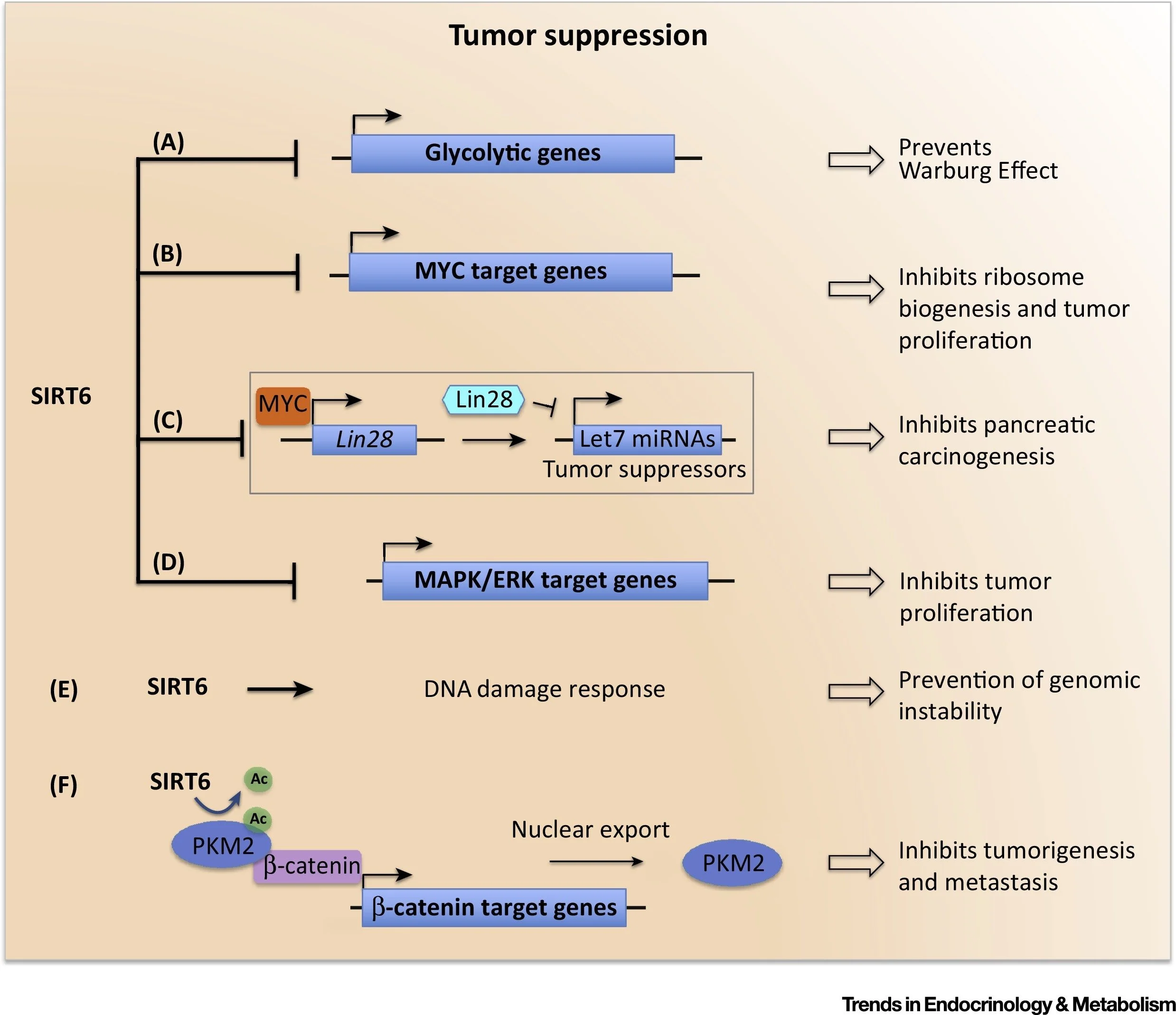
Genome Maintenance: SIRT6 stabilizes DNA-dependent protein kinase at DNA breaks and promotes efficient double-strand break repair
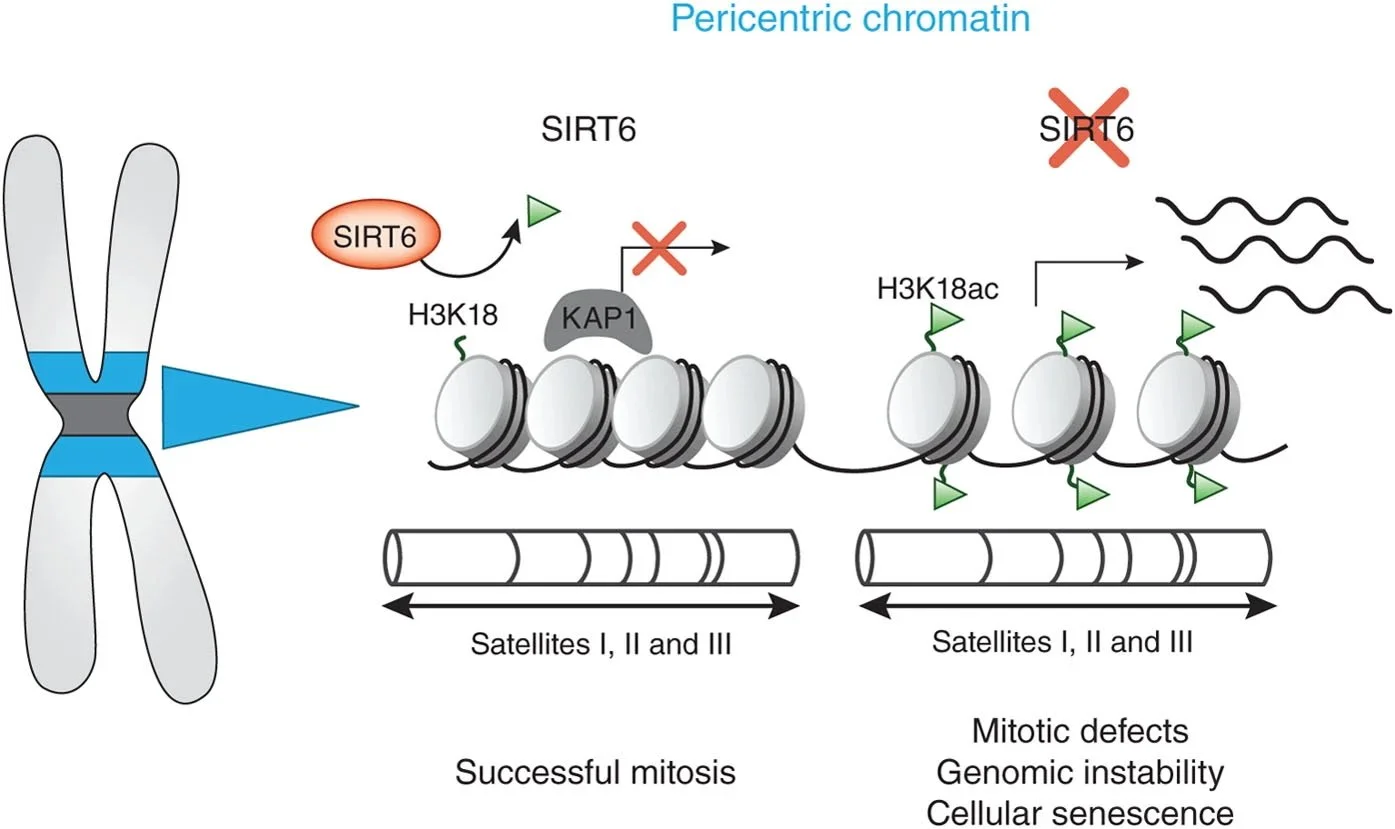
Heterochromatin Silencing: We revealed that SIRT6 maintains pericentric heterochromatin through H3K18 deacetylation, preventing accumulation of pathologic transcripts that trigger cellular senescence
Metabolic Regulation: SIRT6 coordinates metabolic homeostasis through multiple mechanisms, including control of glucose metabolism in a tumor-suppressive manner
Telomere Protection: SIRT6 maintains telomeric chromatin structure and prevents telomere dysfunction, a hallmark of cellular aging
Stress-responsive Regulation: The activity and function of SIRT6 are subject to regulation, for instance, through dynamic phosphorylation on Serine 10 (S10) during mitosis, which is CDK1-dependent and may link SIRT6 functions to mitotic or stress-related signaling.
Physiological Relevance: SIRT6-deficient mice exhibit dramatic premature aging phenotypes, including shortened lifespan, metabolic defects, and genomic instability. Conversely, SIRT6 activation shows promise for extending healthspan and combating age-related diseases.
SIRT7: The Nucleolar Sentinel
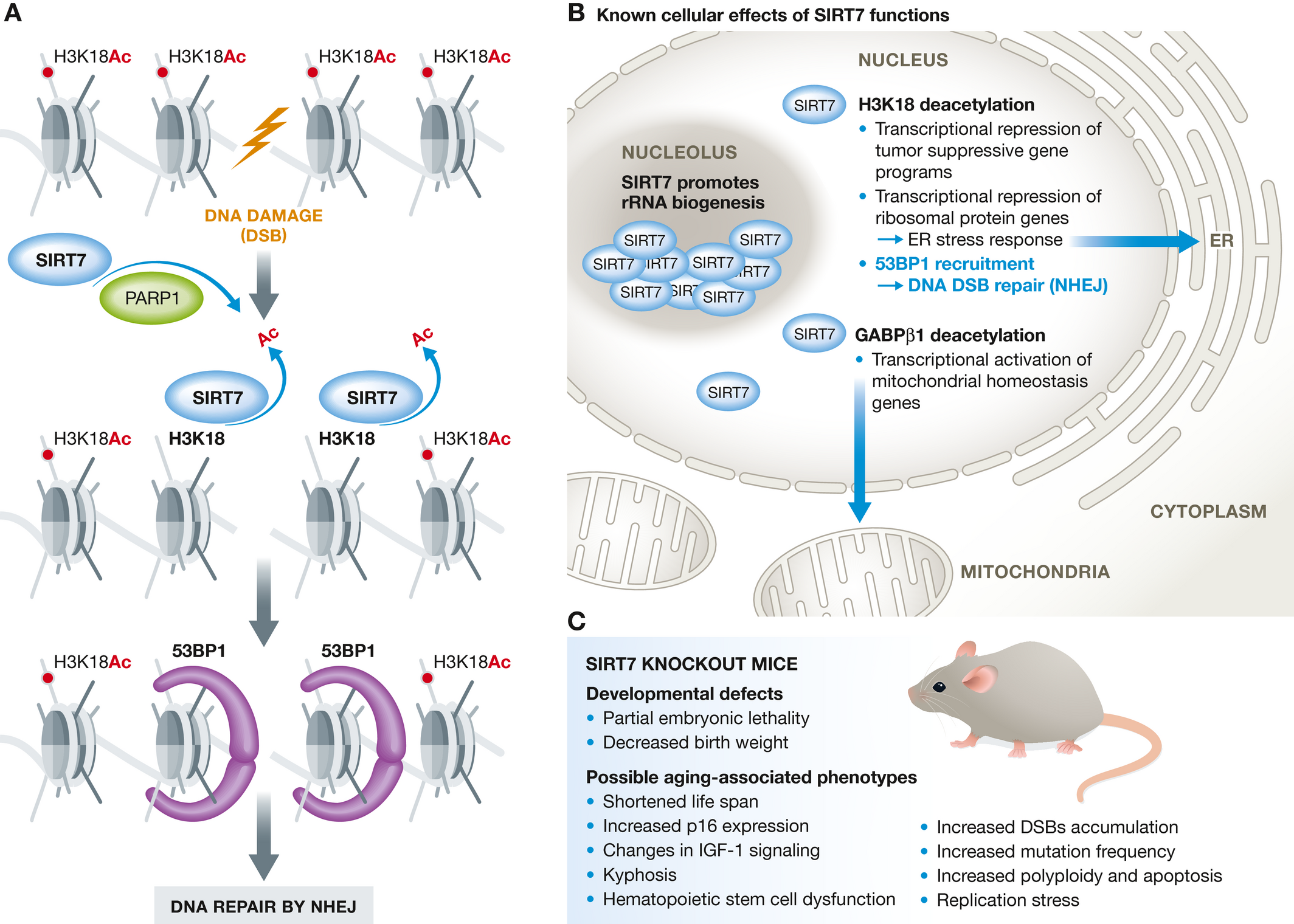
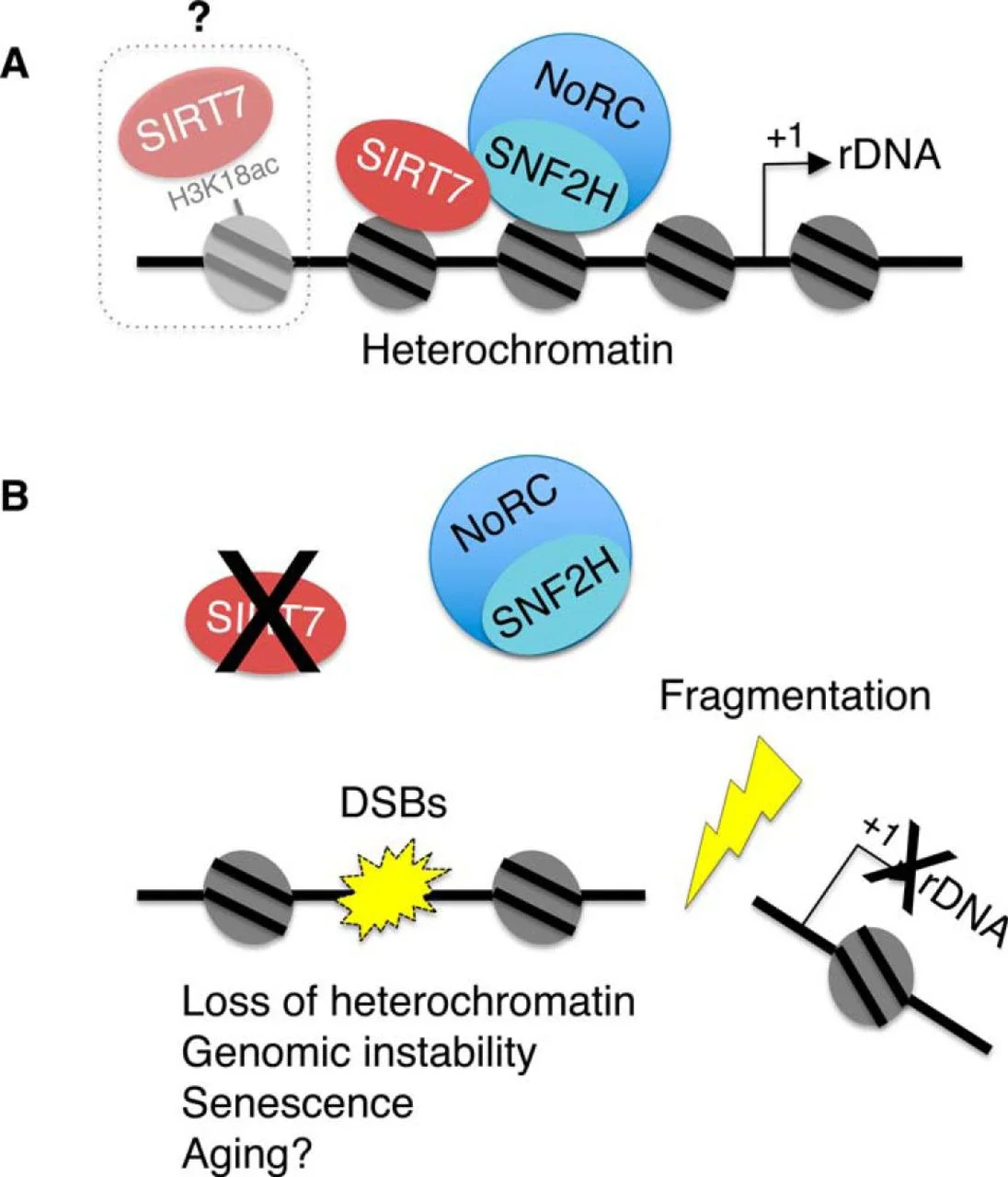
SIRT7 localizes primarily to nucleoli under basal conditions but dynamically shuttles to the nucleoplasm in response to stress. We identified H3K18ac as SIRT7's first substrate and recently discovered its role in deacetylating H3K36ac—a modification with emerging importance in cancer and aging.
Key Functions and Mechanisms:
Oncogenic Transformation: Our work demonstrated that SIRT7 maintains cancer cell identity through H3K18 deacetylation at tumor-suppressive genes
Ribosome Biogenesis: SIRT7 promotes rDNA transcription and rRNA processing
Senescence Prevention: We discovered that SIRT7 guards against a novel senescence pathway triggered by ribosomal DNA instability
Stress Response: SIRT7 responds to metabolic and genotoxic stress by translocating from nucleoli, coordinating cellular adaptation programs by regulating H3K36ac levels at stress-responsive genes throughout the nucleus.
Physiological Relevance: SIRT7 levels correlate with cancer aggressiveness and poor prognosis. Remarkably, SIRT7 depletion can reverse oncogenic transformation, while in normal cells, SIRT7 protects against aging-associated dysfunction. Ongoing work in the lab has also identified a profound role for SIRT7 in metabolic homeostasis.
Want to learn more? Reviews from the lab:
M Ibanez, KF Chua. Chromatin and Nuclear Signaling: SIRT7 Function in the Nucleolus and Beyond. 2018. Introductory Review on Sirtuins in Biology, Aging, and Disease. [PDF]
L Tasselli, W Zheng, and KF Chua. SIRT6: Novel Mechanisms and Links to Aging and Disease. 2017. Trends in Endocrinology & Metabolism [PDF]
S Paredes and KF Chua. SIRT7 clears the way for DNA repair. 2016. The EMBO Journal. [PDF]
E Fang, M Scheibye-Knudsen, KF Chua, M Mattson, D Croteau, and V Bohr. Nuclear DNA damage signalling to mitochondria in ageing. 2016. Nature Reviews Molecular Cell Biology [PDF]
L Tasselli and KF Chua. Methylation gets into rhythm with NAD+-SIRT1. 2015. Nature Structure and Molecular Biology [PDF]
S Paredes, L Villanova and KF Chua. Molecular Pathways: Emerging Roles of Mammalian Sirtuin SIRT7 in Cancer. 2014. Clinical Cancer Research [PDF]
L Tasselli and KF Chua. Metabolism in the driver’s seat. 2012. Nature [PDF]
Some of our recent publications:
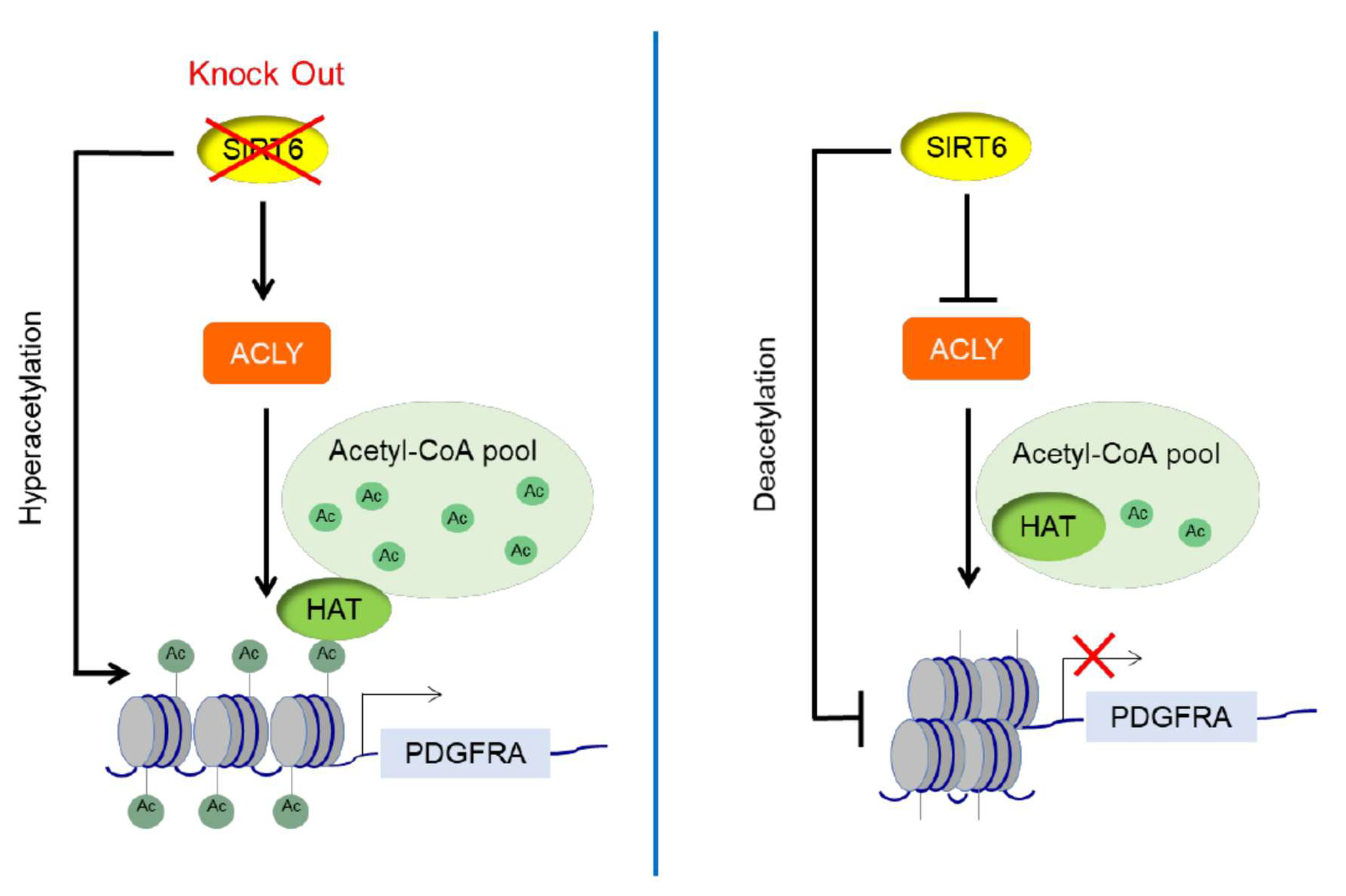
Zheng et al., MDPI 2021: Mammalian SIRT6 Represses Invasive Cancer Cell Phenotypes through ATP Citrate Lyase (ACLY)-Dependent Histone Acetylation [PDF]
Model for the interplay between SIRT6 and ACLY, with a feed-forward loop to repress gene expression by coordinately regulating both histone deacetylation and availability of acetyl-CoA for histone acetylation.
A novel chemical biology approach reveals H3K36ac as a novel substrate for SIRT7.
SIRT7-dependent rDNA silencing via recruitment of SNF2H preserves genomic stability and protects from aging.
Tasselli et al., NSMB 2016: SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence [PDF]
Acting as a H3K18 deacetylase, SIRT6 retains KAP1 on chromatin, thus repressing spurious transcription from heterochromatic satellite repeats, which in turn protects against genomic instability.